RESEARCH
Molecular phylogenetics, population genetics, and comparative genomics
 Current research examines historical patterns of diversification among genes, genomes, and organisms. Various research projects use molecular characters to understanding evolution at several levels of organization including molecular evolution at the genetic or genomic level, divergence of populations and species, and phylogenetic patterns among species and higher taxa. Taxonomically, we are interested in all fishes with particular emphasis on North American freshwater fishes; however, several projects involve other animal groups such as amphipods, mussels and frogs. We are also interested in application of high performance computing to analysis of molecular data, and methods of phylogenetic inference. Current research examines historical patterns of diversification among genes, genomes, and organisms. Various research projects use molecular characters to understanding evolution at several levels of organization including molecular evolution at the genetic or genomic level, divergence of populations and species, and phylogenetic patterns among species and higher taxa. Taxonomically, we are interested in all fishes with particular emphasis on North American freshwater fishes; however, several projects involve other animal groups such as amphipods, mussels and frogs. We are also interested in application of high performance computing to analysis of molecular data, and methods of phylogenetic inference.
Fish Tree of Life
What are phylogenetic relationships among the >30,000 species of bony fishes? We are actively investigating this question (see DeepFin). A major project is the Euteleost Tree of Life. Euteleostei is the crown group of ray-finned fishes, comprising some 346 families, 2,935 genera, and 17,419 species at present count (Nelson, 2006). Over one third of all vertebrate species are euteleosts, including virtually all of the economically important fishes and important genomic, developmental, and physiological model systems. The EToL project is directed at understanding the unparalleled morphological, genomic, developmental, physiological, and behavioral diversity of euteleost fishes by reconstructing a well-resolved and well-supported "backbone" phylogeny of the group.
RECENT NEWS! New Tree of Life and a Revised Classification of Bony Fishes
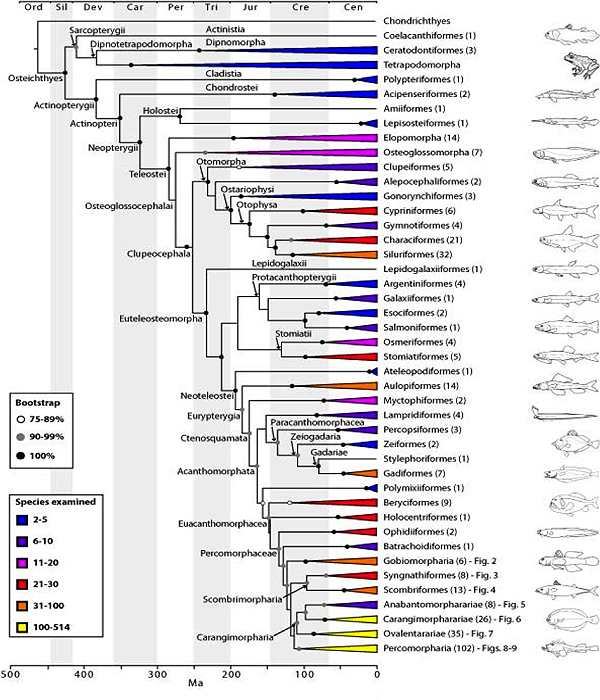
The ETOL team has produced the first comprehensive phylogeny of fishes based on DNA sequence data from 21 genes for 1416 species. The molecular phylogeny is time-calibrated using 59 fossil calibration points and forms the basis for a revised classification. See our recent papers Betancur et al. 2013 and Broughton et al. 2013.
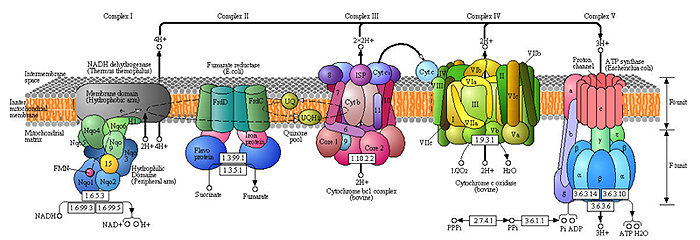
Evolutionary Genomics of the Oxidative Phosphorylation System
OXPHOS produces the majority of ATP used by eukaryotic cells. This complex system involves over 70 proteins encoded by both the mitochondrial and nuclear genomes. We are interested in the evolutionary dynamics resulting from very different characteristics of the two genomes and how this affects interactions among subunits and OXPHOS efficiency. In particular, we are investigating the complex history of natural selection acting on OXPHOS genes from organisms with different metabolic ATP demands.
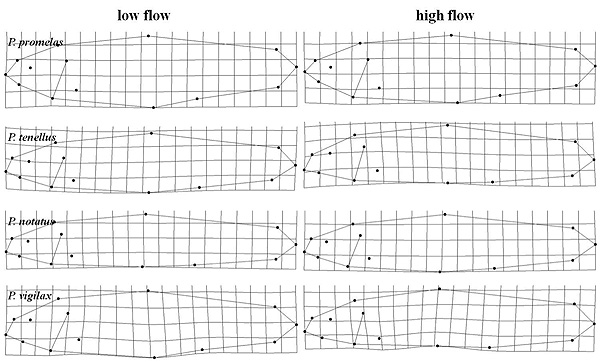
Environmental Genomics
We are exploring how genomic variation and patterns of gene expression are related to environmental variation. This includes responses to environmental change in different species and what attributes of invasive species make them successful competitors outside their native ranges. Of particular interest are the genetic mechanisms that control phenotypic plasticity and how these can contribute to speciation.
Patterns of Nucleotide Change and Character Quality in Phylogenetic Analysis
 This involves assessing the relative historical information content of characters that conflict on phylogenetic trees (homoplasy). We would like to know, when is homoplasy misleading, when is homoplasy not misleading, and how do we recognize the difference and make use of this information? This involves assessing the relative historical information content of characters that conflict on phylogenetic trees (homoplasy). We would like to know, when is homoplasy misleading, when is homoplasy not misleading, and how do we recognize the difference and make use of this information?
Genealogical Patterns
We use genealogical patterns to understand processes of population divergence, hybridization, and the nature of reproductive barriers. What do gene genealogies reveal about the historical, demographic, and selective factors associated with speciation in natural populations? A focal taxon is fishes in the genus Cyprinella.
Systematics, Biogeography, and Conservation of Fishes
We put an emphasis on species native to central and southwestern North America. This includes molecular population genetic and phylogenetic studies of native species and field surveys of the distribution and abundance of fishes in Oklahoma. |

